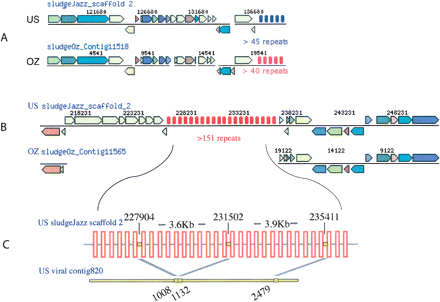
Sample alignments of homologous regions in the dominant US and OZ strains showing substitution (A) and insertion (B) of CRISPR elements. CRISPR repeats are indicated by sets of vertical bars with colors denoting different repeat sequences. Total number of repeats for each CRISPR element is unknown because of incomplete sequence information in the draft assembly; therefore, a minimum estimate is given. (C) Schematic magnification of dominant CAP CRISPR element and a contig from the phage virion metagenome revealing a phage that has previously infected CAP. All spacers targeting the phage had the same orientation. The starting positions of each spacer and the matching segments in the phage are indicated. The drawing is not to scale, and the actual number of repeats is significantly higher.











