Table 5.
Switch error rates estimated from fully sequenced clones
Click on table to view larger version.
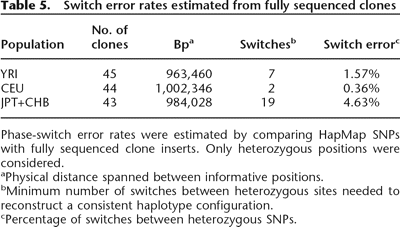
Phase-switch error rates were estimated by comparing HapMap SNPs with fully sequenced clone inserts. Only heterozygous positions were considered.
aPhysical distance spanned between informative positions.
bMinimum number of switches between heterozygous sites needed to reconstruct a consistent haplotype configuration.
cPercentage of switches between heterozygous SNPs.











