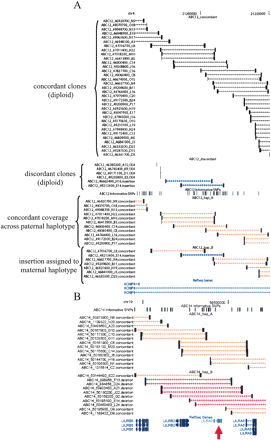
Interactive browser of mapped fosmid clones. Screenshots from an interactive browser (http://hgsv.washington.edu) displaying mapping information for clones from eight fosmid libraries are depicted. (A) The complete concordant clone coverage (black bars) along with two putative insertion clones (blue bars) indicates that ABC12 is heterozygous for an insertion in an intron of the KCNIP4 gene. The presence of four, mapped one-end anchored clones suggests that the insertion may involve sequence not represented elsewhere in the genome assembly. Haplotype sorting of clones from this region indicates that the insertion allele is on the maternal haplotype (haplotype B). The names and classifications of all clones are given on the left side of the image. (B) A deletion impacting the LILRA3 gene is assigned to the nontransmitted haplotype of sample ABC14 (red arrow). A study of a German cohort suggests that deletions of this locus may be associated with risk of multiple sclerosis (Koch et al. 2005). The deletion is known to exist at a high frequency in Japanese populations where haplotypes from this locus form two distinct clades (Hirayasu et al. 2006). The ability to obtain contiguous sequence from haplotypes carrying the deletion and insertion configuration may clarify the role of this locus in human evolution and disease.











