Summary of SFL partner prediction for ADA2
Click on table to view larger version.
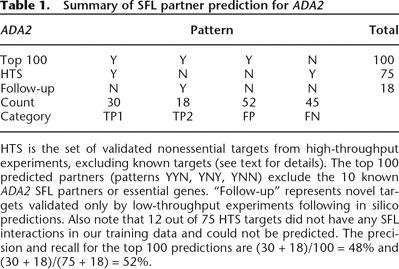
HTS is the set of validated nonessential targets from high-throughput experiments, excluding known targets (see text for details). The top 100 predicted partners (patterns YYN, YNY, YNN) exclude the 10 known ADA2 SFL partners or essential genes. “Follow-up” represents novel targets validated only by low-throughput experiments following in silico predictions. Also note that 12 out of 75 HTS targets did not have any SFL interactions in our training data and could not be predicted. The precision and recall for the top 100 predictions are (30 + 18)/100 = 48% and (30 + 18)/(75 + 18) = 52%.











