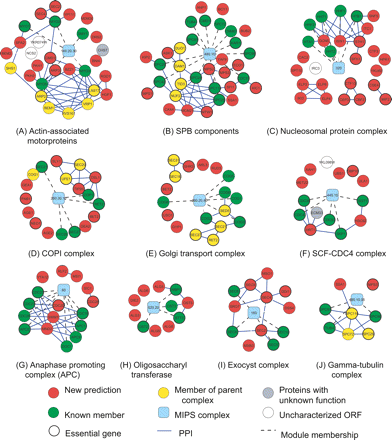
Modules from complex-based search. Cyan nodes (module) are labeled with the ID of the MIPS complex used as query. Known members of the query complex with no SFL interaction in our training set are not shown. (Blue edges) Physical interactions absent from MIPS but present in high-throughput data (Gavin et al. 2006; Krogan et al. 2006; Stark et al. 2006). Physical interactions between known members are not shown.











