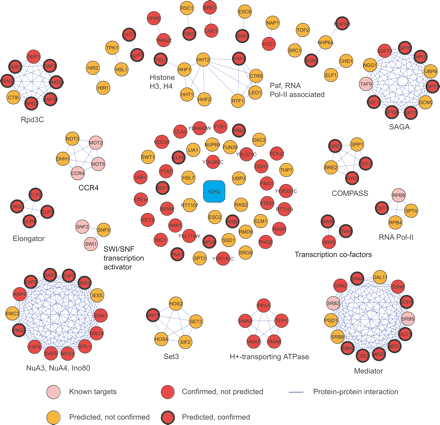
SFL targets of ADA2 were obtained by combining high-throughput screening results and predictions by the odd-parity kernel G−, validated by random spore analysis or tetrad dissection. (Confirmed) Experimentally tested positive. Protein–protein interactions are derived from the MIPS database and Krogan et al. (2006) and Gavin et al. (2006). The SFL targets of ADA2 are most enriched of histone and chromatin modification complexes and the mRNA transcription machinery.











