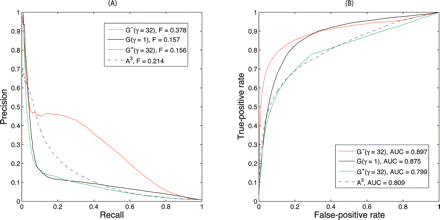
Figure 3.
Performance of predicting genetic interactions from BioGRID by three diffusion kernels and the raw counts of length-3 paths. The optimal diffusion parameters used are γ = 32 for G−, γ = 1 for G and γ = 32 for G+. The raw counts of length-3 paths is A3. The odd-parity diffusion kernel G− significantly outperforms all other methods. (A) Precision-recall curves. (B) Receiver operating characteristic curves.











