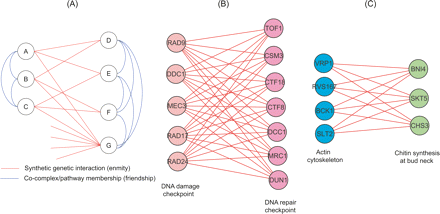
The parallel pathway model in the synthetic genetic interaction network. (A) A synthetic genetic interaction network is analogous to a social network of enemies. The two branches represent genes in parallel pathways, analogous to people in competing social groups. Nodes within the same branch are friends (or members of the same pathway), while nodes in different branches are enemies (or genes with a synthetic lethal genetic interaction). (B) The functionally overlapping DNA damage checkpoint genes and DNA repair checkpoint genes have enriched between-pathway SFL interactions (Pan et al. 2006). (C) Two groups of cell cortex genes have enriched between-pathway SFL interactions (Kelley and Ideker 2005).











