Flanking gene method data for 24 Arabidopsis gene categories documenting cases of recent transposition
Click on table to view larger version.
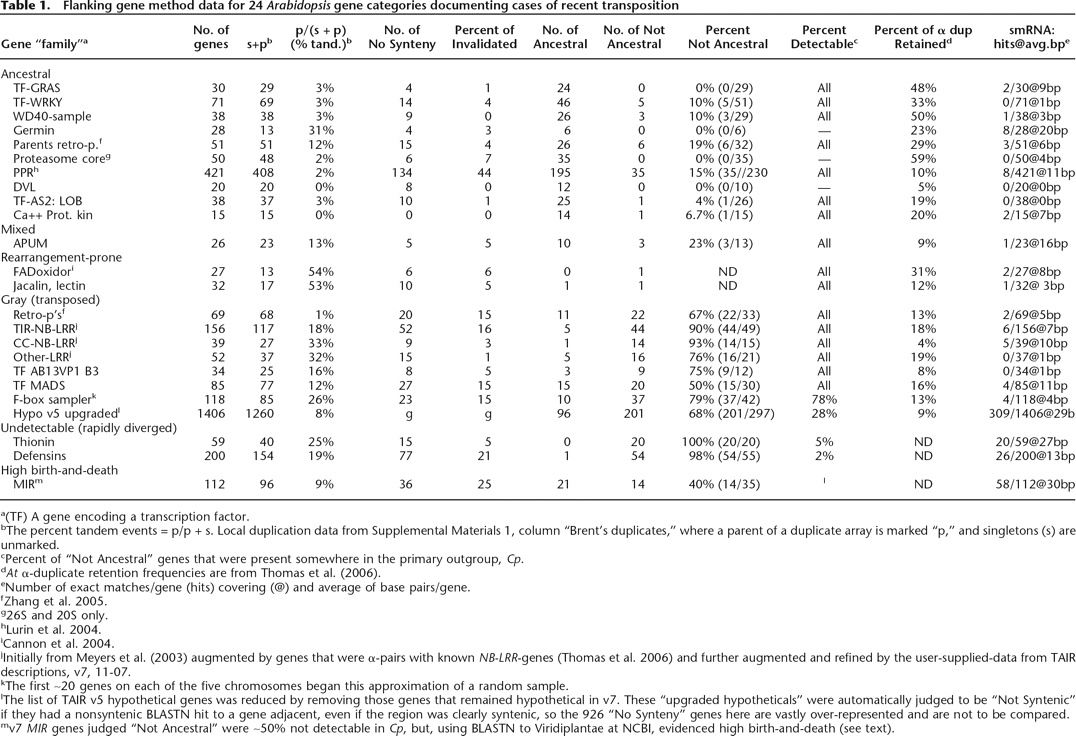
a(TF) A gene encoding a transcription factor.
bThe percent tandem events = p/p + s. Local duplication data from Supplemental Materials 1, column “Brent’s duplicates,” where a parent of a duplicate array is marked “p,” and singletons (s) are unmarked.
cPercent of “Not Ancestral” genes that were present somewhere in the primary outgroup, Cp.
dAt α-duplicate retention frequencies are from Thomas et al. (2006).
eNumber of exact matches/gene (hits) covering (@) and average of base pairs/gene.
fZhang et al. 2005.
g26S and 20S only.
hLurin et al. 2004.
iCannon et al. 2004.
jInitially from Meyers et al. (2003) augmented by genes that were α-pairs with known NB-LRR-genes (Thomas et al. 2006) and further augmented and refined by the user-supplied-data from TAIR descriptions, v7, 11-07.
kThe first ∼20 genes on each of the five chromosomes began this approximation of a random sample.
lThe list of TAIR v5 hypothetical genes was reduced by removing those genes that remained hypothetical in v7. These “upgraded hypotheticals” were automatically judged to be “Not Syntenic” if they had a nonsyntenic BLASTN hit to a gene adjacent, even if the region was clearly syntenic, so the 926 “No Synteny” genes here are vastly over-represented and are not to be compared.
mv7 MIR genes judged “Not Ancestral” were ∼50% not detectable in Cp, but, using BLASTN to Viridiplantae at NCBI, evidenced high birth-and-death (see text).











