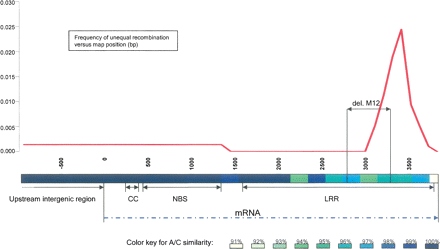
Distribution of unequal recombination events along the consensus sequence of the NBS–LRR paralogs A and C (3737 bp) and their upstream noncoding regions (870 bp). Similarity between paralogs A and C is shown using a color-coded bar below the distribution graph. Seven of the 11 A–C recombinants and the single B/C recombinant (M1–M7, M13) were localized in a highly variable segment of the LRR region. Recombination frequency in this region was significantly higher (P < 0.0005) than the overall frequency measured along the complete gene. A 468-bp internal deletion in paralog B of M12 overlaps with the A/C recombination hot-spot, suggesting that an imprecisely repaired double-strand break also occurred in the high-recombination region.











