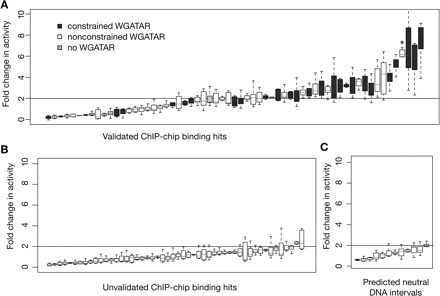
Activities of DNA segments occupied by GATA1 in enhancer assays. (A) The results of eight to 24 determinations of enhancer activity after transfection for each occupied DNA interval added to a luciferase expression plasmid, rank-ordered by activity. The distribution of results for each occupied segment is represented as a box plot, with the internal line indicating the median, the box extending to the first and third quartiles, and the whiskers extending to the most extreme data point that is no more than 1.5 times the interquartile range. Boxes for occupied segments with at least one constrained GATA1 binding motif (i.e., preserved outside rodents) are shaded dark gray, those with nonconstrained motifs (i.e., found only in rodents) are white, and those with no match to a binding site motif shaded light gray. The asterisk marks the activity for DNA segment GHP304. (B) The results of enhancer activity for DNA intervals that were positive for occupancy by ChIP-chip (many passed only a low stringency threshold, see Supplemental material) but were not validated by qPCR. (C) The transfection results for DNA segments predicted to be neutral because they were not positive for ChIP-chip data. Intervals that generate at least a twofold increase in activity compared with that of the parental reporter gene plasmid (corresponding to the mean of the negative controls plus 3 SDs) in at least two repeats of the assay are considered to be active as enhancers.











