Table 1.
A comparison of the observed and expected substitution distances for the hold-one-out cross-validation experiments
Click on table to view larger version.
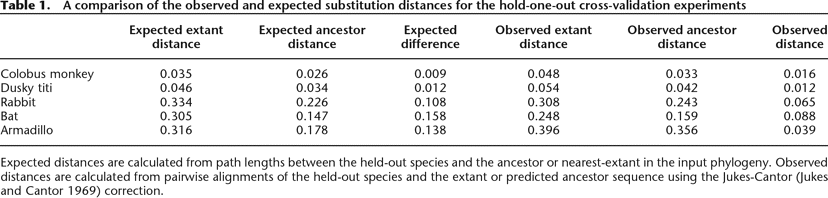
Expected distances are calculated from path lengths between the held-out species and the ancestor or nearest-extant in the input phylogeny. Observed distances are calculated from pairwise alignments of the held-out species and the extant or predicted ancestor sequence using the Jukes-Cantor (Jukes and Cantor 1969) correction.











