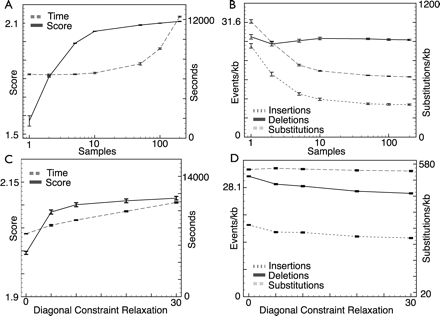
The effects of sample rate and constraint relaxation on time, likelihood, and mutation rates. The effects of changing the sample rate and diagonal constraint relaxation on (A,C) the log-likelihood (arbitrary scale), computation time and (B,D) rates of insertions (of up to 10 bases), deletions (of up to 10 bases), and substitutions for the ancestor reconstruction of the CFTR region. Results are the average of five runs; error bars represent the maximum variation (plus or minus) observed. Runs were performed on Xeon (Pentium 4) processors with 2.4-GHz clock speeds and 4 GB of memory. The sample rates examined were 1, 2, 5, 10, 50, 100 (default), and 200 samples. Log scales are shown for the sample rate x-axis. Diagonal constraint relaxations of 0, 5, 10 (default), 15, 20, and 30 were used. All other parameters were set at their default values.











