Data showing an assessment of a set of projected exon chains
Click on table to view larger version.
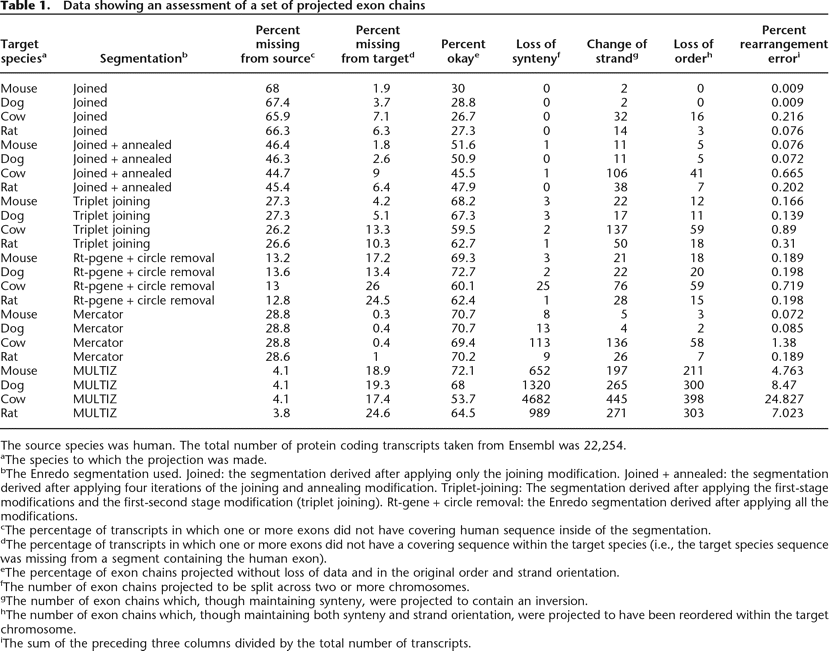
The source species was human. The total number of protein coding transcripts taken from Ensembl was 22,254.
aThe species to which the projection was made.
bThe Enredo segmentation used. Joined: the segmentation derived after applying only the joining modification. Joined + annealed: the segmentation derived after applying four iterations of the joining and annealing modification. Triplet-joining: The segmentation derived after applying the first-stage modifications and the first-second stage modification (triplet joining). Rt-gene + circle removal: the Enredo segmentation derived after applying all the modifications.
cThe percentage of transcripts in which one or more exons did not have covering human sequence inside of the segmentation.
dThe percentage of transcripts in which one or more exons did not have a covering sequence within the target species (i.e., the target species sequence was missing from a segment containing the human exon).
eThe percentage of exon chains projected without loss of data and in the original order and strand orientation.
fThe number of exon chains projected to be split across two or more chromosomes.
gThe number of exon chains which, though maintaining synteny, were projected to contain an inversion.
hThe number of exon chains which, though maintaining both synteny and strand orientation, were projected to have been reordered within the target chromosome.
iThe sum of the preceding three columns divided by the total number of transcripts.











