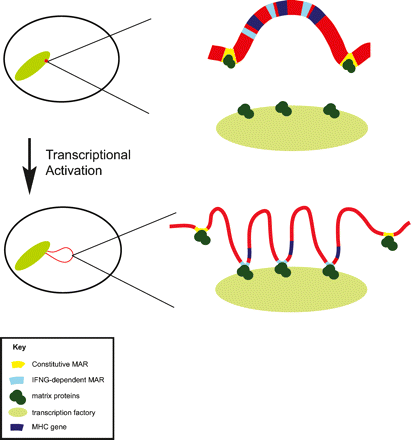
Model showing dynamic changes in chromatin organization of the MHC locus upon transcriptional up-regulation. In cells that are not expressing MHC class II genes, such as uninduced fibroblasts, the 4 Mb MHC locus (red) is positioned within the chromosome 6 territory anchored by constitutive MARs (yellow) to nuclear matrix proteins (dark green), forming stable chromatin loops. Upon up-regulation of gene expression the entire MHC locus adopts a more open, decondensed chromatin structure, represented by the thinning and lengthening of the chromatin line, and the entire MHC region is repositioned outside of the chromosome territory. IFNG-dependent MARs (light blue) are recruited to the nuclear matrix, folding chromatin into smaller loops and facilitating the interaction of chromatin and transcription factors (light green). Many IFNG-dependent MARs are positioned immediately adjacent to genes. The constitutive MARs remain attached to nuclear matrix proteins following transcriptional up-regulation, and some may have an insulatory function.











