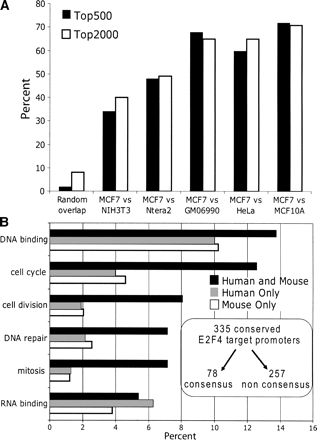
Conserved E2F4 targets are more highly enriched in classical E2F target gene categories, but are not enriched in consensus E2F motifs. (A) The percentage of promoters that were identified in either the top 500 or top 2000 sets of ranked promoters from both the mouse 3T3 and human MCF7 E2F4 ChIP-chip experiments was determined. For comparison, the percentage expected by random chance and the percentage overlap of E2F4 targets from MCF7 cells with E2F4 targets from four other human cell lines is also shown. (B) Functional annotations were performed as described in Figure 4. The top categories are shown for the sets of E2F4 target promoters identified only in the duplicate human E2F4 MCF7 ChIP-chip experiments (794 promoters), only in the duplicate E2F4 3T3ChIP-chip experiments (977 promoters), or in all four human and mouse E2F4 ChIP-chip experiments (335 promoters). The x-axis indicates the percentage of each set represented by the different categories; see Supplemental Table S3B for the P-values for each category. Shown in the inset is the number of the human and mouse conserved E2F4 target promoters that contain or lack a consensus E2F motif.











