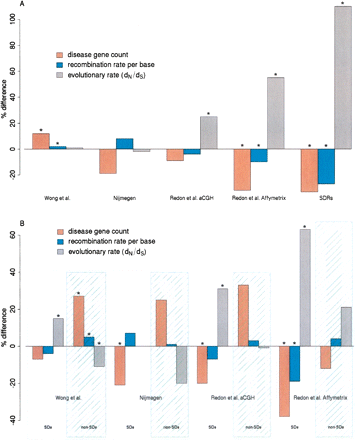
The departures from expected values for evolutionary rates, recombination rates, and disease gene count estimated within Nijmegen CNVRs, Redon et al. Affymetrix CNVRs, Redon et al. aCGH CNVRs, Wong et al. CNVRs and SDRs. Evolutionary rates and disease gene counts have been obtained only for genes that entirely lie within these CNVRs. Results are shown as percentage differences from values expected from random sampling of the human genome reference assembly (see Methods). (A) Departures of the rate of protein evolution (dN/dS), recombination rate per base (cM/bp) and disease gene count are shown as percentage differences from the value expected from random sampling of the human genome. Bars annotated with an asterisk (*) are significant at P < 0.025. (B) Departures of the same properties as in A, after splitting the different CNVR data sets according to their overlap with SDs.











