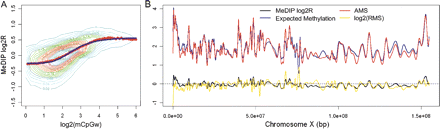
MeDIP logR is a logistic function of the log-methylation level. (A) The logistic model (blue line) describes the association between MeDIP log2R and the log2 observed methylation level. Using fully methylated genomic DNA, we determined the methylation level based on the weighted count of mCpG in a 1 kb window centered at each probe (mCpGw, using chromosome X tiling array). The red dots identify the median MeDIP logR within each bin across the entire mCpGw dynamic range. (B) The absolute methylation score (AMS) and expected DNA methylation level across the X chromosome. The model parameters estimated from the first hybridization have been applied to the MeDIP logR derived from the second hybridization (black line) to determine the log2(AMS) (red line). Log2(mCpGw) corresponds to the expected methylation level (blue line). The log relative methylation score (RMS) is drawn with the gold line. All the estimates are chromosome-wide smoothed by means of cubic splines.











