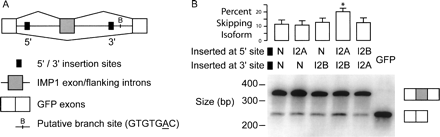
I2A/B motif pairs can suppress splicing of an intervening exon. (A) Mini-gene construct for interrogating I2A/B motif pair, constructed by inserting exon 12 of the human IGF2BP1 gene and its flanking introns into the middle of the ORF in an eGFP expression construct. (B) The I2A/B motif pair promotes exon skipping in HeLa cells. The five indicated constructs containing I2A, I2B, or neutral (N) motifs inserted near the 5′ss or 3′ss were transfected into HeLa cells. Twenty-four hours later RNA was extracted and semi-quantitative RT-PCR using primers targeted to reporter exons 1 and 3 was performed to assay for relative isoform levels. (Top) Quantization of skipping levels. Data shown are mean + SEM for eight replicates—two PCRs for each of four transfection experiments. *I2A/B motif pair shows significantly more skipping than N/N control (P = 1.1 × 10−5 by one-sided t-test; 1.75-fold increase in skipping). At the 5% level, none of the other skipping levels is significantly greater than that of N/N. (Bottom) Representative gel showing levels of inclusion isoform (upper band) and skipping isoform (lower band). Last lane, intronless GFP control.











