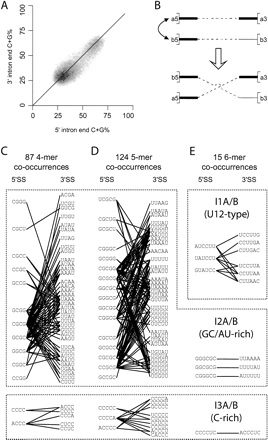
coCOA detects three clusters of motif pairs that co-occur at 5′ and 3′ ends of human introns. (A) G+C content in the first 80 nt (x-axis) and last 80 nt (y-axis) of introns is correlated. A density plot of intron co-GC content is shown for a set of 53,326 constitutive human introns, with the darker/lighter squares corresponding to higher/lower intron density, respectively. The diagonal line y = x is shown for reference. (B) co-GC shuffling. (Above) Two hypothetical introns, A and B, with 5′/3′ ends a5/a3 and b5/b3. Intron A has high G+C content at both ends (thick lines). Intron B has high G+C content at the 5′ end, but lower G+C content near the 3′ end (thin solid line). Since the introns have similar G+C content at their 5′ ends, these ends can be swapped. (Below) Co-GC shuffled introns. The beginning of intron B (b5) is now paired with the end of intron A (a3), and the beginning of intron A (a5) is now paired with the end of intron B (b3). Overall co-GC content of the set of introns is preserved. (C–E) Preferentially co-occurring k-mer pairs detected by coCOA are shown for k = 4, 5, and 6 at P ≤ 4−2k, corresponding to a single expected false positive for each value of k. In each panel, k-mers occurring in the first 80 nt of introns are shown at left under “5′SS”; those occurring in the last 80 nt are shown at right under “3′SS”. The co-occurrences could all be grouped into three clusters, denoted I1, I2, and I3, with the 5′ss and 3′ss motifs designated A and B, respectively.











