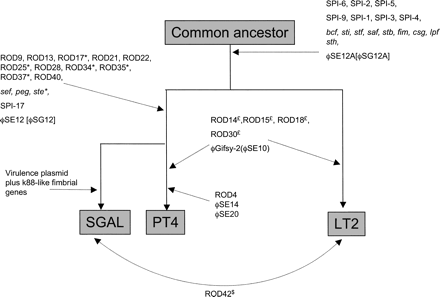
Line diagram to represent the whole-genome differences of S. Enteritidis PT4, S. Typhimurium strain LT2, and S. Gallinarum strain 287/91. A summary of the observed loss and gain of RODs described in Table 2. The diagram is based on the assumption that following the divergence of PT4 and LT2 from a common ancestor PT4 and GAL have subsequently diverged. Branches are not intended to infer phylogenetic distance. Evidence that a locus was once present in LT2, PT4, or GAL (see legend to Figure 1) but has subsequently been deleted from that genome is marked by the suffix *, $, or £, respectively. Brackets indicate the name for locus in GAL. Parentheses indicate the name for locus in PT4. Dotted arrows mark the position in the pseudo-tree at which that ROD(s) appears











