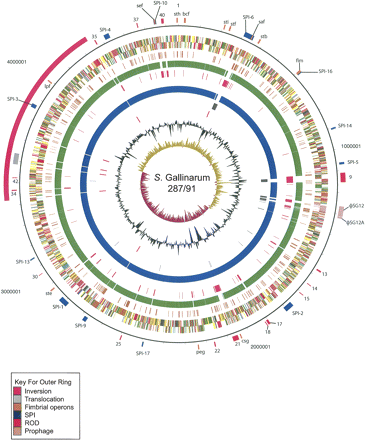
Circular representation of the S. Gallinarum 287/91 chromosome. For a full description of this figure, see legend to Figure 2. The exception is that circle 5 shows the position of S. Gallinarum 287/91 pseudogenes. Circles 6 and 8 show the position of S. Gallinarum 287/91 genes which have orthologs (by reciprocal FASTA analysis) in S. Typhimurium LT2 (all CDS colored green) or S. Enteritidis PT4 (all CDS colored blue), respectively. Circles 7 and 9 show the position of S. Gallinarum 287/91 genes that lack orthologs in (by reciprocal FASTA analysis) in S. Typhimurium strain LT2 (all CDS colored pink) and S. Enteritidis PT4 (all CDS colored gray), respectively. Circle 10 shows the position of the S. Gallinarum 287/91 rRNA operons (red).











