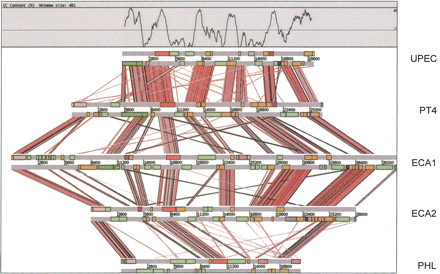
Comparison of the ROD21 locus of S. Enteritidis with related genomic islands. ACT comparison (http://www.sanger.ac.uk/Software/ACT) of amino acid matches between the complete six-frame translations (computed using TBLASTX) of ROD21 compared with related loci uropathogenic E. coli strain CFT073 (UPEC), Erwinia carotovora sbsp. atroseptica strain SCRI1043 (two loci: ECA1 and ECA2), and Photorhabidus luminescens,sbsp. laumondii TT01 (PHL; see Methods). The red bars spanning between the genomes represent individual TBLASTX matches. CDS are marked as colored boxes positioned on the horizontal gray DNA bars: (orange) genes conserved in two or more of the genomic islands; (light green) variable genes of unknown function; (dark pink) hnsB; (dark blue) hnsT; (light pink) integrase; (dark green) type IV pilin-associated genes; (red) plasmid-related mobility functions; (salmon pink) transposase-related genes; (yellow) tRNA genes; (light blue)repeats. The G + C profile for the UPEC loci is shown above. The scale is marked in base pairs.











