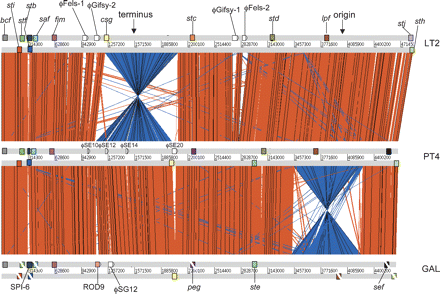
Global comparison between S. Typhimurium, S. Enteritidis, and S. Gallinarum. ACT comparison (http://www.sanger.ac.uk/Software/ACT) of amino acid matches between the complete six-frame translations (computed using TBLASTX) of the whole-genome sequences of S. Typhimurium LT2 (LT2), S. Enteritidis PT4 (PT4), and S. Gallinarum 287/91 (GAL). Forward and reverse strands of DNA are shown for each genome (light gray horizontal bars). The red bars between the DNA lines represent individual TBLASTX matches, with inverted matches colored blue. The position of all the fimbrial operons in these three genomes are marked as colored boxes positioned on the forward and reverse strands of DNA. Analogous fimbrial operons are colored the same. The boxes of fimbrial operons that include pseudogenes are crossed with a white line. Other genomic features are only shown if they constitute breaks in synteny between genomes. The position of the origin and terminus are marked (solid black arrows).











