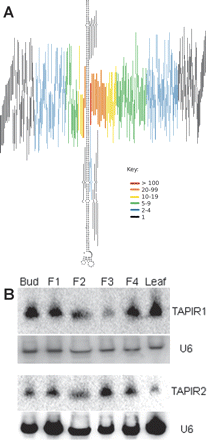
TAPIR-derived sRNAs. (A) Predicted secondary structure of one particular TAPIR element with lines representing the sRNA sequences mapping to the two arms of the hairpin. The color of the lines specifies the abundance of the sequences in the library. (B) Northern blot shows the accumulation of the two most abundant, overlapping sRNAs (exact positions are shown in Supplemental Fig. 6) from TAPIR elements. Membranes were stripped and reprobed for U6 to show equal loading. F1, 1–3 mm; F2, 5–7 mm; F3, 7–11 mm; F4, 11–14 mm.











