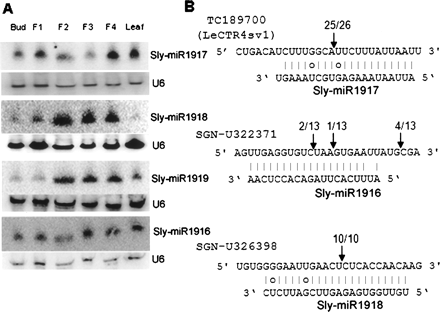
Expression and target validation of new nonconserved tomato miRNAs. (A) Northern blot analysis of new miRNAs showed that sly-miR1918 and sly-miR1919 accumulate preferentially in the fruit. The U6 probe was used to show equal loading. Different size fruits were analyzed: F1, 1–3 mm; F2, 5–7 mm; F3, 7–11 mm; F4, 11–14 mm. (B) The result of target validation for three new miRNAs. (Arrows) The 5′ ends of cleavage products mapped inside the displayed sequence. Target EST sequences are shown on top of the miRNA sequences.











