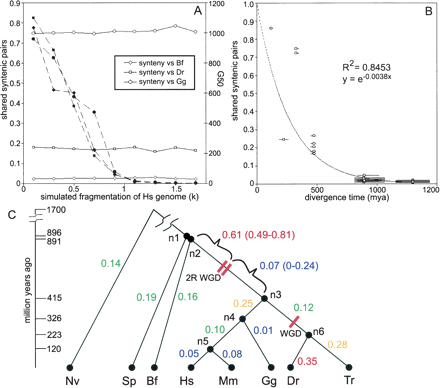
Rates of synteny loss throughout vertebrates and their ancestors. (A) Estimates of conserved synteny are robust to genome fragmentation. The human genome was artificially fragmented into scaffolds of random lengths according to a Pareto distribution where k satisfies the equation, scaffold size = 1/U1/k, and U is a random number between 0 and 1. These fragmented human genomes were then compared to amphioxus (Bf), chicken (Gg), or zebrafish (Dr). As k increases, the G50 size of the fragmented human genome decreases (dashed lines), but the syntenic shared pair metric remains relatively consistent (solid lines). G50 is the gene number such that 50% of the assembled genome lies in scaffolds containing at least G50 genes. (B) Conserved synteny was measured between all pairwise combinations of human, fugu, zebrafish, chicken, mouse, amphioxus, sea urchin, and sea anemone and then plotted relative to the divergence age of the comparison. The values are well fit by an exponential curve. (C) Syntenic distances were apportioned to the known species tree and then divided by the estimated evolutionary time in each branch to obtain rates of synteny loss. Internal nodes are labeled n1–n6. The highest rates of loss are observed in the period after the vertebrate divergence from amphioxus but before the early vertebrate WGD events (n2–2R WGD), and in the terminal zebrafish lineage (n6–Dr). Species abbreviations are human (Hs), mouse (Mm), chicken (Gg), fugu (Tr), zebrafish (Dr), amphioxus (Bf), sea urchin (Sp), sea anemone (Nv).











