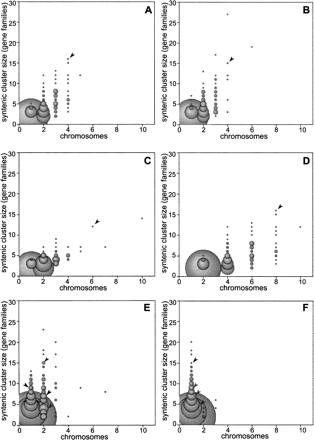
Synteny group distributions reveal vertebrate duplication events. The size of each syntenic group, measured by the number of gene families in the group, is plotted against the number of chromosomes over which the group is spread. The bubble sizes are proportional to the number syntenic groups at each point. (A–C) Vertebrate synteny groups built by comparison to amphioxus: (A) human, (B) chicken, and (C) zebrafish. In A and B, the largest groups, with the most conserved synteny, are present on four chromosomes, and a steady reduction in chromosome coverage is seen as the group size decreases. (C) The zebrafish groups are spread out past four chromosomes. (D) As a control, the chromosome coverage of the human groups shown in A was doubled, creating a simulation of a new WGD event on top of the early chordate duplications. This plot shows a similar chromosome spread to C, and post-WGD gene loss in zebrafish could account for the sparser plot. (E) Zebrafish synteny groups, built by comparison to the human genome, show that the largest synteny groups cover two to three chromosomes. (F) Human synteny groups, built by comparison to the zebrafish genome, show a strong peak at one chromosome, as expected in the absence of WGD events. Arrowheads within the plots indicate the bubbles that contain the Hox clusters. In comparisons between amphioxus and vertebrates (A–D), the Hox genes form a single synteny group, while, in comparisons between fish and tetrapods (E–F), they subdivide into four separate groups, indicating that the cluster duplicated twice within the early vertebrate lineage.











