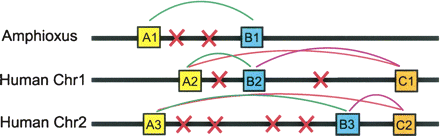
Identifying and grouping pairs of syntenic genes. Syntenic gene pairs were identified in three steps. First, genes from the two genomes are grouped into orthologous families (A, B, and C). Next, both genomes are searched for syntenic “family combinations”—pairs of genes from two ortholog families that are found in close proximity in more than one genomic location. Genes are in close proximity if they have no more than 10 intervening genes (shown here as red x’s). In this example, three syntenic family combinations are identified (A–B, A–C, and B–C). The A–B combination is present in both amphioxus and humans, while the A–C and B–C combinations are only present in humans. These syntenic gene pairs can then be merged to generate segments of syntenic genes (A1–B1, A2–B2–C1, and A3–B3–C2). In the human genome these syntenic segments form a “synteny group,” which contains three gene families and is present on two chromosomes.











