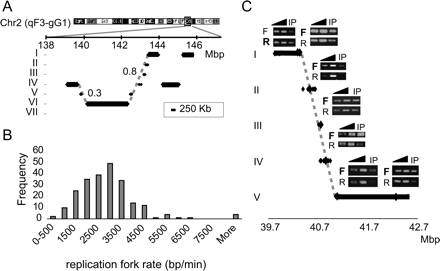
Large replicons. (A) Large replication segments (>250 kb; black bars) and small replication segments (<250 kb; each flanked by two dots) are shown for a small region in chromosome 2. The vertical position of each line represents its assigned ToR, and the horizontal position represents its location on chromosome 2. Dashed gray lines represent the predicted replication timing of the region assuming a movement of a single replication fork from the early to the late region. The numbers next to the dashed lines represent the RMSD values between the dots and the line. Note the good agreement between the predicted replication time (dashed line) and the actual timing (black dots). Additional examples of such regions are shown in Supplemental Figure S7. (B) Histogram of the replication fork rate, as deduced from the analysis of regions containing large replicons. (C) Replication fork direction was analyzed by measuring the relative amounts of the two strands in BrdU-labeled cells and treated with emetine, which inhibits lagging strand synthesis (see Methods). The ToR of a region on chromosome 1 is shown as in A. The regions for which replication fork direction was analyzed are marked by large diamonds. For each such region, linear PCR with either a forward (F) or reverse (R) primer, followed by semi-quantitative PCR was performed (two dilutions of total DNA [triangles] and emetine-resistant BrdU-labeled DNA [IP]). The direction of the replication fork can be deduced from the relative amount of the reverse and forward reactions. Note that all five regions assayed along the diagonal line show the same direction of the fork (marked by bold letters), strongly suggesting that this entire region is replicated by a single fork.











