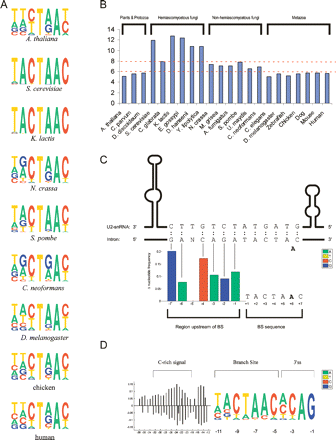
Comparative analysis of the branch site. (A) Sample BS motifs, for selected organisms (see Supplemental Fig. S4 for all organisms). (B) Information content of the 7-nt BS, calculated as in Fig. 2. (C) Upstream extension of the BS signal in certain fungi. The region upstream of the BS in each intron was aligned, and positional bias plots of this region were formed. Results are shown for the seven positions immediately upstream of the BS in E. gossypii. Each of the significantly enriched (P < 0.05) nucleotides in this region is complementary to nucleotides in U2 snRNA directly downstream to the nucleotides that bind the BS sequence. The U2 snRNA nucleotides lie in a potential unpaired region, between two U2 snRNA stem-and-loop I and IIa structures (Ares 1986). The bold “A” nucleotide marks the BS nucleotide. (D) The 3′ intron end in Y. lipolytica. An alignment of the last 11 intronic positions is presented in the form of a pictogram. This alignment uses the 3′ss as anchor. The BS is clearly apparent, between positions −11 and −5. A C-rich signal upstream of the BS is presented in the form of positional bias plots.











