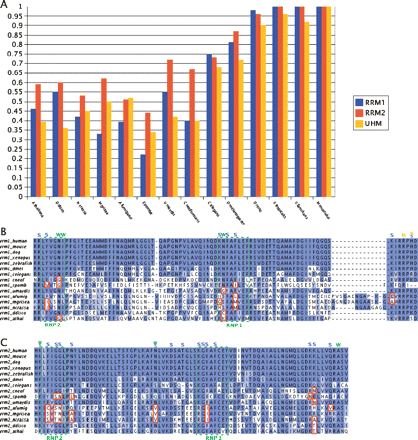
Comparative analysis of the RRM domains between U2AF2 homologs: (A) The conservation levels of the RRM domains of each organism are compared with their human counterpart. Xenopus tropicalis was used instead of chicken (see Methods). In B and C a conservation analysis of key residues in the RRM1 and RRM2 domains, respectively, is presented. These residues have been shown to be required for PPT binding in human (Sickmier et al. 2006). Residues labeled by “W” interact with the RNA through a water molecule, whereas residues labeled by “S” and “M” interact through the side-chain and main-chain, respectively. The dashed green boxes highlight the RNP1 and RNP2 motifs and the red boxes mark substitutions, with respect to metazoans, that change the biochemical properties in key residues that are responsible for RNA binding. In fungi, the following substitutions, with respect to human, can be observed: aromatic residues (F and Y) by nonaromatic ones (V, I, L, M, C, S) in the RNP1 and RNP2 regions of both RRMs. In the RRM1, polar to nonpolar substitutions: K to G at RNP1 and N to G in the RNP1 and RNP2; and polar basic (K) to nonpolar neutral (A) and polar acidic (E) at the C-terminal region (panel B). Similarly, in RRM2 of fungi we observed substitutions of a nonpolar residue (G) to polar ones (T and S) in the RNP2; polar to nonpolar substitutions: N to V, L, A near the RNP motifs, and Q to V at the C-terminal region; and basic to neutral (K to N, A) at the C-terminal region. In the RRM1 of A. thaliana there is a polar (N) to nonpolar (G) substitution in the RNP2 and a neutral (N) to basic (K) substitution in the RNP1. In the RRM1 of D. discoideum there is a substitution N to C, which reduces the polarity at this position. Finally, in the RRM2 of both species, there is a basic (K) to neutral (T) substitution in the C-terminal region.











