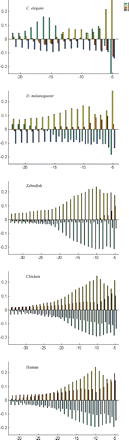
PPT regions in metazoans. The PPT region was defined as the region between the BS and the 3′ss. For each organism, the median distance between the termination of the BS sequence and the 3′ss was determined, and positional bias plots of the region between this position and position −5 relative to the 3′ss are shown. Briefly, positional bias plots show the bias of each nucleotide at each position relative to the background frequency, by showing the difference (Δ) between the position-specific and the background frequency (see Methods). The y-axis presents the extent and nature of the bias, with positive and negative values denoting nucleotides that appear more, and less, than expected, respectively. Note: Adenosines, thymidines, cytosines, and guanines are visualized in green, yellow, red, and blue, respectively. Note: The bars at position −5 of C. elegans are truncated: The bias for T at this position is 0.54, while that of A is −0.27. See Supplemental Fig. S4 for results of other organisms.











