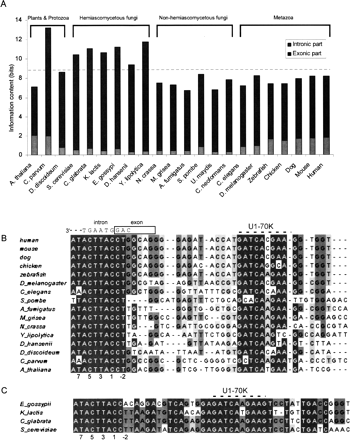
(A) Information content (measured in bits) of the 5′ss sequence for each of the organisms. Information content is shown separately for the 4-nt exonic part and the 8-nt intronic part (marked in gray and black, respectively). Total information content is the sum of the two. (B) Multiple sequence alignments of the 5′ region of the U1 snRNA molecule in metazoans, protozoans, plant, and non–S. cerevisiae-like fungi. The scheme at the top shows a sample 5′ss containing the human consensus nucleotides, opposite the corresponding positions on U1 snRNA, and the location of the 5′ss positions relative to the exon/intron boundary is marked at the bottom. The binding site of the splicing factor U1–70K is marked as well. (C) Multiple sequence alignment of the 5′ region of U1 snRNA in S. cerevisiae-like fungi, which are characterized by a longer U1 snRNA sequence and by a different secondary structure (Kretzner et al. 1990). The 5′ss positions and the U1–70K binding site are marked as in B.











