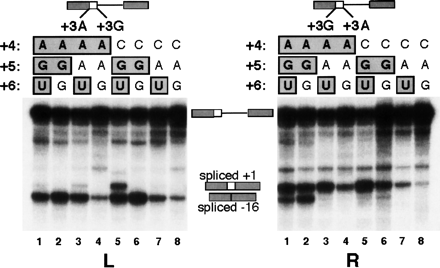
The deleterious effect of the +3 A-to-G mutation can be alleviated by correcting positions +4 and/or +5. In this competition assay, we created two sequences from the ACADSB exon 3 5′ss: one with +3A and another with +3G (for the experimental strategy, see Fig. 5; for the ACADSB 5′ss sequences, see Fig. 6). The same mutations were simultaneously introduced at positions +4, +5, and +6 at both 5′ss, to test their compensatory effects vis-à-vis the +3 A-to-G mutations. The nucleotides at positions +4, +5, and +6 are listed above the lanes, and the consensus nucleotides are boxed. The left and right panels are the same, except that the positions of the +3A and +3G 5′ss are swapped (indicated on the top diagram). With a few exceptions, the general trend is that the +3G 5′ss was used to a greater extent as the number of consensus nucleotides at positions +4 to +6 was increased (lanes 1–3, 5). The degree of use of the +3G 5′ss was not the same for identical mutants in the two panels (for details, see text), but despite these positional effects, the trend remains. The +3G 5′ss can be activated by correcting position +4 alone (lane 3) or +5 alone (lane 5). The rescue by +5 alone is surprising and indicates that associations between nonadjacent positions can affect splice-site efficiency.











