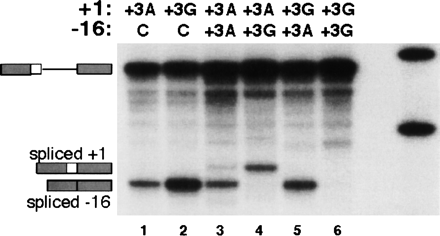
The +3 A-to G-mutation in the ACADSB 5′ss has a severe effect on splicing efficiency. By using the competition strategy described in Figure 5, we performed three experiments. First, the ACADSB wild-type 5′ss (+3A, GGG/GUACAU) and the mutant (+3G, GGG/GUGCAU), which differs by having a G at +3, were placed in competition against the beta-globin cryptic (GUG/GUGAGG) 5′ss at −16 (Treisman et al. 1983). The cryptic 5′ss at −16 was the only 5′ss used (lanes 1,2), indicating that both +3A and +3G 5′ss are very weak. Second, a competition was done between +3A and +3G, which resulted in splicing only via the +3A 5′ss (lanes 4,5). Third, the same 5′ss was placed on the same substrate at both locations. Whereas the substrate with two +3A 5′ss resulted in use of both −16 and +1 5′ss (lane 3), two +3G 5′ss did not show any splicing (lane 6). Together, these results indicate that the +3A 5′ss is much stronger than the +3G 5′ss.











