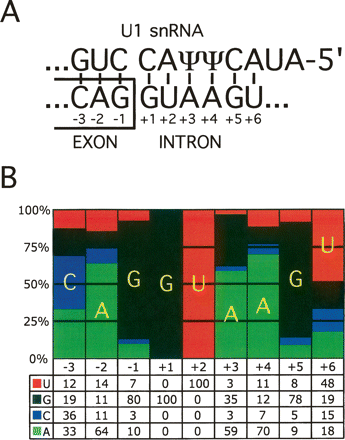
Features of the 5′ splice site (5′ss). (A) Base pairing between the 5′ end of the U1 snRNA and the consensus 5′ss sequence. ψ denotes pseudo-uridine, which is a modified uridine nucleotide capable of base pairing to both A and G nucleotides. The conventional numbering of positions relative to the exon–intron boundary is indicated. For example, in the text −1G refers to nucleotide G at position −1. (B) A pictorial representation of the position weight matrix (PWM) of the human U2-type GT-AG 5′ss; the bars represent the percentages of nucleotides at each position of the 5′ss. The actual percentages are shown below the representation.











