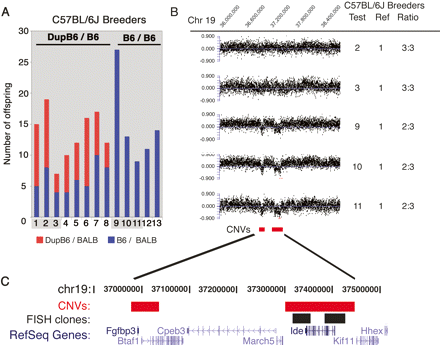
Breeding and CGH confirm CNV between individual C57BL/6J breeders. (A) The number of F1 offspring in each genotype class is shown for 13 different C57BL/6J breeders. Eight breeders (1–8) are heterozygous at the locus (DupB6/B6) and transmitted an extra copy of the Ide locus to ∼50% of their offspring. Five breeders (breeders 9–13) are presumed homozygous for the locus (B6/B6) and did not transmit a duplicated copy of the locus. Shading highlights C57BL/6J breeders where copy number was confirmed by CGH analysis. (B) CGH detects CNVs in the C57BL/6J strain. Data are shown for hybridizations of five different C57BL/6J breeders each compared to a sixth C57BL/6J breeder as a reference sample. The normalized log2 ratio is plotted for each probe within the portion of chromosome 19 indicated. Test and reference sample numbers correspond to the animal numbers for individual C57BL/6J breeders shown in panel A. For each pair of samples the copy number ratio is given to indicate if hybridizations revealed an equal genomic copy number (3:3) or a reduced copy number (2:3) between the test and reference sample. (C) The location of the CNV and fosmid clones used for FISH are shown relative to RefSeq genes in the region.











