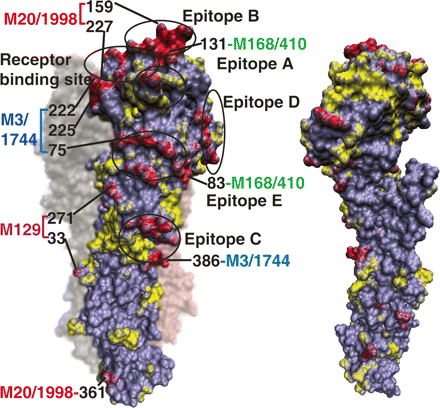
Module-based H3N2 amino acid substitutions mapped onto the HA structure. Amino acid substitutions from 1968 to 2006 are mapped onto the (left panel) exposed and (right panel) buried surface of the HA monomer. Residues are colored by module-based amino acid changes (red), non-module-based changes (yellow), and intact residues (purple). The five antibody epitopes (Wiley et al. 1981) and the receptor-binding site (Skehel and Wiley 2000) are circled. The module-based amino acid substitutions carrying two or more residues in 1998–2006 are labeled.











