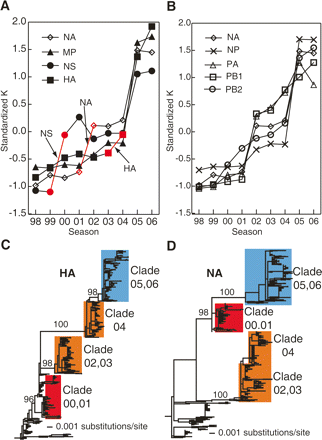
Evolutionary patterns of individual influenza gene segments. (A,B) Average standardized connectivity K of H3N2 nucleotide co-occurrence networks for gene segments (A) HA, NA, MP, and NS; and (B) NA, NP, PA, PB1, and PB2 from 1998–2006. The largest changes in connectivity K in HA, NA, and NS before 2004 are labeled in red and indicated by arrows. (C,D) Phylogenetic trees of (C) HA and (D) NA nucleotide sequences from influenza A viruses sampled from 1997 to 2006. Bootstrap values are shown for the key nodes. The clades for strains during 2000–2006 were labeled using the season from which the majority of the strains were isolated.











