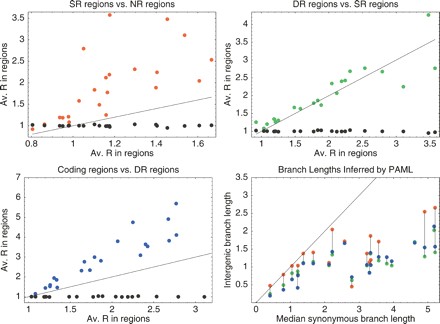
Comparison of average R values in different regions for 22 clades of bacteria. The red dots in the top-left panel show the average R in SR regions (vertical axis) against the average R in NR regions (horizontal axis). The green dots in the top-right panel show the average R in DR regions (vertical) against the average R in SR regions (horizontal). The blue dots in the bottom-left panel show the average R in coding positions (vertical) against the average R in DR regions (horizontal). The black dots in all panels show the average R in silent positions (vertical). The line y = x is also shown in all panels. The bottom-right panel shows, for each clade, the total branch lengths in the phylogenetic trees as inferred by PAML on alignment columns from NR (red), SR (green), and DR (blue) regions, as a function of the total branch length in the phylogenetic tree inferred from the silent positions (horizontal). Dots corresponding to the same clade are connected by vertical lines.











