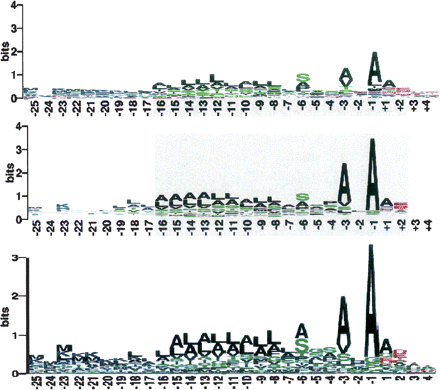
Figure 11.
(Top) Sequence logo for the amino acid sequence motif of all signal peptides identified by MS/MS analysis. Position −1 correspond to the last residue of the signal peptide. (Middle) Sequence logo for Gram-negative bacteria employed by PrediSi (Hiller et al. 2004). (Bottom) Sequence logo for Gram-negative bacteria employed by SignalP (Nielsen et al. 1997).











