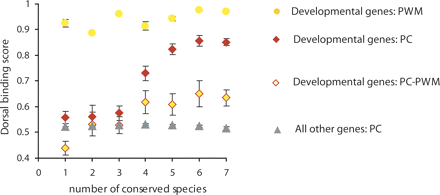
PC, but not PWM, method of binding site scoring detects the relationship between the binding site sequence conservation and its binding affinity to Dorsal. The Rel binding sites in enhancer regions of developmental genes described by Papatsenko and Levine (2005) were defined by either the principal coordinate (PC) analysis (red diamonds) or position-weight-matrix (PWM) analysis as in Ref (Papatsenko and Levine 2005) (orange circles). The relationship between the sequence conservation of Rel binding sites and their average binding affinity to Dorsal is shown. The sites defined by the PC analysis and not overlapping with the PWM are shown in red diamonds filled with orange (PC–PWM); the sites defined by the PC analysis in front of other genes are shown in gray triangles.











