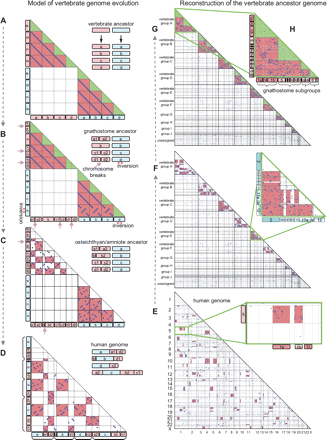
Model of vertebrate genome evolution and reconstruction of the ancestral genome. (A) For simplicity, suppose that the ancestral chromosome had 10 genes. The 2R WGD produced ohnologs (blue dots along the diagonal line in the triangular dot plot) in the duplicated chromosomes. (B) Chromosome breaks and inversions may have altered the order of ohnologs on the sister chromosomes. (C) In the course of early vertebrate genome evolution, the ancestral gene order was disrupted by many inversions, resulting in scattered ohnolog dots. (D) Eventually, CVL blocks were distributed across several human chromosomes by intensive interchromosomal rearrangements. (a–d) A typical model of genome evolution involving the 2R WGD. In the next step, we handle real human genome data. (E) This is a real instance of the dot plot in D. CVL blocks were ordered from the human chromosomes 1 to X, and ohnologs shared among these CVL blocks were plotted. (Red) Regions representing pairs of paralogous CVL blocks with a great number of ohnologs (P < 10−4, see Methods). (F) This corresponds to the state in C. CVL blocks were reordered in such a way that paralogous CVL blocks were grouped so that each group represented one ancestral vertebrate chromosome (see Methods). (G) This state corresponds to that in B. CVL blocks within individual vertebrate groups were further reordered to obtain ancestral gnathostome subgroups (namely, chromosomes), which were duplicated from a single ancestral vertebrate chromosome by the 2R WGD events. The partition of subgroups that optimizes the significance defined in the Methods. Rectangles and triangles are colored in accordance with those in B to make the correspondence clear. (H) The vertebrate group A was decomposed into four gnathostome subgroups by statistical analysis, indicating that the ancestral chromosome underwent 2R WGD.











