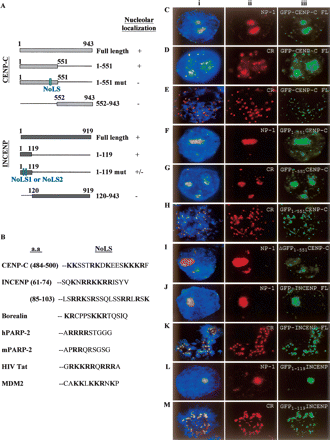
Determination of the nucleolus localization sequence (NoLS) motifs on CENPC1 and INCENP. Full-length and truncated GFP-CENPC1 and GFP-INCENP constructs were transfected into human 293 cells. More than 20 cells from each transfection experiment were scored for their subcellular distribution pattern. (A) Schematic diagrams for GFP fusion constructs for CENPC1 (top panel) and INCENP (bottom panel). Numbers indicate the positions of amino acid residues of the proteins or peptide being expressed. The locations of the NoLS motifs are indicated. Plus and minus signs indicate positive and negative localization to the nucleolus, respectively. (B) Amino acid comparisons between NoLSs of CENPC1, INCENP, and nucleolus proteins borealin, human and mouse PARP2, HIV Tat protein, and MDM2 (Lohrum et al. 2000). Bold letters denote basic amino acids. The amino acid positions of the CENPC1 and INCENP NoLSs are shown. (C–M) Human 293 cells were transfected with either full-length or truncated GFP-CENPC1 and GFP-INCENP constructs (see A). The localization of full-length (FL) GFP-CENPC1 (detected using anti-GFP antibody) (Ciii–Eiii; green) to nucleoli (Ciii), and interphase and metaphase centromeres (Diii,Eiii, respectively), are indicated by coimmunostaining with nucleophosmin-1 (Cii; red/NP-1) and CREST (Dii,Eii; red/CR) antibodies. Some examples of the colocalization of GFP-CENPC1 (Diii,Eiii; green) and CREST at the interphase centromeres (Dii; red) and metaphase centromeres (Eii; red) are presented. Direct visualization of GFP-CENPC1 signals without anti-GFP antibody gave similar results (data not shown). The NoLS at the N-terminal portion of CENPC1 (GFP-CENPC1 amino acids 1–551) efficiently targeted nucleolar (Fiii; green) and interphase and metaphase centromeres (Giii,Hiii, respectively; green) as indicated by costaining with nucleophosmin-1 (Fii; red/NP-1) and CREST (G,Hii; red/CR), respectively. The mutation of Lys residues within NoLS (1–551 mut in A) abrogated the recruitment to both the centromere (not shown) and nucleolus (Iiii; green), as indicated by NP-1 staining (Iii, red). The GFP fusion CENPC1 protein carrying the C-terminal fragment (amino acids 552–943) also fails to target to both the nucleolus and centromere (data not shown). Both full-length (FL) GFP-INCENP (Jiii,Kiii; green) and truncated (amino acids 1–119; containing unmodified NoLS1 and NoLS2) GFP-INCENP (Liii,Miii; green) proteins targeted to the nucleolus and centromere, as indicated by costaining with antibodies against nucleophosmin-1 (Jii,Lii; red/NP-1) and centromere (Kii,Mii; red/CR). Mutating NoLS1 or NoLS2 showed varying effects on nucleolar and centromere localization (see text). The GFP fusion protein carrying the C-terminal portion of INCENP (amino acids 120–943) targeted to neither the nucleolus nor the centromere (data not shown). (i) Merged images of ii and iii.











