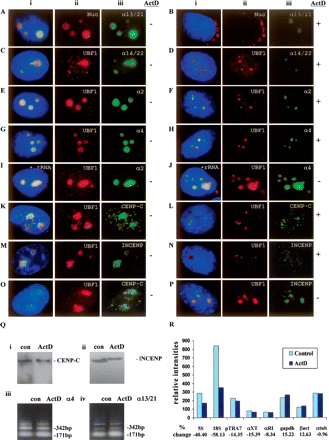
(A–J) Subnuclear distribution of centromeric α-satellite RNA and proteins in HeLa cells. Unless otherwise stated, images were captured on a conventional fluorescence microscope. Simultaneous RNA-FISH detection of α-satellite RNA specific for chromosomes 13/21 (Aiii; green), 14/22 (Ciii; green), 2 (Eiii; green) and 4 (Giii; green); and immunofluorescence detection of nucleolar proteins nucleolin (Nuc) (Aii; red) and UBF1 (Cii,Eii,Gii; red) in human HeLa cells. ActD treatment at 0.05 μg/mL resulted in the delocalization of α13/21- (Biii; green), α14/22- (Diii; green), α2- (Fiii; green), and α4- (Hiii; green) satellite RNA, nucleolin (Bii; red), and UBF1 (Dii,Fii,Hii; red) from the nucleoli. UBF1 displayed a typical relocalization toward the nucleolar periphery to give a concentrated cap-like signal at the tip of the nucleoli (Dii,Fii,Hii; red). Competition with tRNA and rRNA (10× excess amount) did not affect the detection of α-satellite RNA at the nucleoli as exemplified by the chromosome 2- and 4-specific α-satellite RNA signals (Iiii,Jiii; green). (K–N) Nucleolar accumulation of CENPC1 (Kiii; green) and INCENP (Miii; green), as indicated by coimmunostaining with UBF1 (Kii, Mii, respectively; red). CENPC1 and INCENP both delocalized from the nucleoli following ActD treatment (Liii,Niii, respectively; green). (O) Confocal laser scanning microscopy analysis showing nucleolar accumulation of CENPC1 by differential interference contrast images (iii; green) and coimmunostaining with UBF1 (ii; red). (P) Confocal microscopy analysis showing nucleolar accumulation of INCENP (iii; green) and coimmunostaining with UBF1 (ii; red). (i) Merged images of ii and iii. Antibodies used were as described in the Methods, except for the anti-CENPC1 antibody used in Oiii, which was kindly provided by Dr. Bill Earnshaw. (Q) Effect of ActD treatment on the expression level of CENPC1, INCENP, and α-satellite RNA. Total cell lysates were prepared from HeLa cells with (ActD) and without (con) prior treatment with 0.05 μg/mL ActD for 6 h, and subjected to Western blotting analysis. The expression levels of both CENPC1 (i) and INCENP (ii) remained unchanged following ActD treatment. (iii) Total RNA was prepared from untreated and ActD-treated HeLa cells. RT-PCR was performed on the RNA using primer sets corresponding to the α4- and α13/21-specific α-satellite sequences. A 171-bp ladder of monomeric α-satellite sequences was seen. No significant difference in the intensities of PCR products was detected following 6 h ActD treatment. (R) Nuclear run-on transcription assay to assess the effect of ActD treatment on α-satellite transcription. Equal numbers of nuclei were used for each assay, as determined by the DNA concentration. The effect of ActD on the transcription of α-satellite was measured by comparing the hybridization signals of in vitro-synthesized 32P-labeled run-on transcripts with immobilized DNA. The radioactive signals for the ribosomal genes 5S and 18S were decreased following ActD treatment, but the signals for the transcription of RNA polymerase II-driven genes (such as GAPDH, beta-actin, and alpha-tubulin) and those of α-satellite DNA (pTRA7 [chromosomes 13/14/21-specific], αXT [chromosomes 14/22-specific], and αRI [chromosomes 13/21-specific]) (Lo et al. 1999) were not significantly affected.











