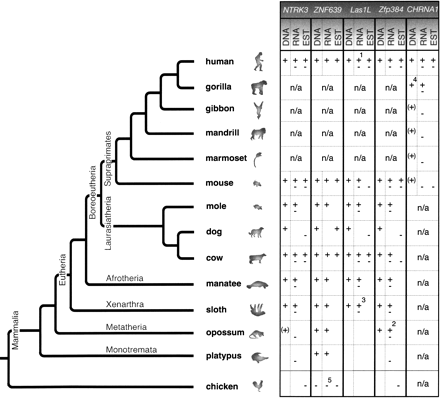
Phylogenetic tree of investigated mammals showing the species distributions of the five internal MIR exonizations. In DNA, “+” indicates the presence of the intronic MIR element leaving the splice sites and reading frame intact, “(+)” its presence, without leaving the reading frame intact, and “−” its absence. In RNA, “+” indicates the presence of the exonized MIR sequence in cDNA derived by RT-PCR, and “−” the presence of a cDNA that does not include the exonized MIR sequence. Both symbols indicate that both splice forms are expressed. EST, expressed sequence tag, “+” including or “−” without the exonized MIR region, available at GenBank (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi). Empty boxes indicate that the corresponding sequence information is not available or not PCR amplifiable; n/a, not analyzed. 1A third splice variant exists, derived from the human MegaMan Human Transcriptome Library (Stratagene). 2Five splice variants exist, three of which include the exonized MIR element. 3RT-PCR performed in the nine-banded armadillo instead of sloth. 4Not shown are “+” symbols under the DNA for chimpanzee and orangutan. 5Unrelated exonized intronic sequence in chicken and a corresponding exonization in the ostrich.











