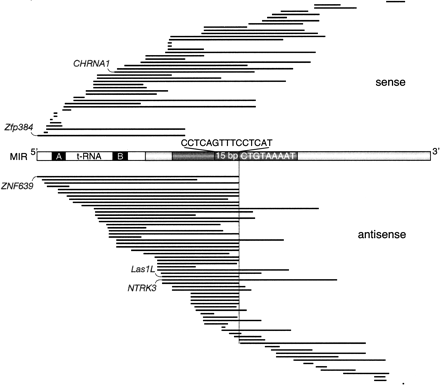
Distribution and orientation of exonized MIR sequences located in protein-coding sequences (CDS). The 107 human exonized MIR sequence regions are aligned against a schematic of a MIR consensus element. The tRNA-related part, including the internal promoter A and B boxes (black), is shown without shading (white). The region shown in dark gray comprises the 70 nt conserved central domain including a highly conserved 15-bp core sequence. The location of the MIR internal, antisense cryptic splice site AG is indicated on the sense strands (CT) by a vertical line. The 9-nt natural 3′ MIR splice site (5′-ATTTTACAG-3′) is shown as the inverse consensus sequence (Supplemental Fig. S3). The exonized MIR regions are represented as black lines; intronic or untranslated region (UTR) portions of the MIRs are not shown. The five experimentally analyzed examples are indicated (CHRNA1, Zfp384, ZNF639, LAS1L, and NTRK3).











