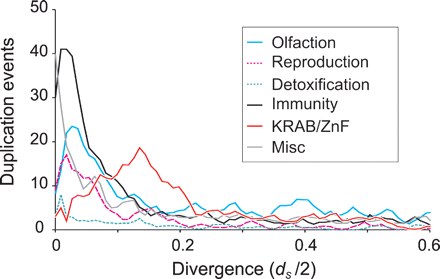
Inferred rates of duplication for Monodelphis inparalogs among different functional categories. The timing of duplication events for Monodelphis-specific inparalogs were inferred from the dS-based rooted phylogenetic tree, and thus appears on a scale of dS/2 units. Although dS values vary substantially among genes (Fig. 3B), on average, a dS/2 value of 0.25 represents ∼90 million years. Genes in most functional classes (including olfaction, reproduction, detoxification, and immunity) follow a profile typical of other mammals with duplications peaking very recently. This would be in accordance with a rapid gene birth and death model. It is likely that reduction in the number of most recent duplications (dS/2 < 0.02) is due partly to the collapse of nearly identical sequence in the assembly, and partly to underprediction of sequence-similar adjacent paralogs and overestimation of very small dS values. The duplications of KRAB-ZnF genes are prominent in peaking at dS/2 ∼ 0.14.











