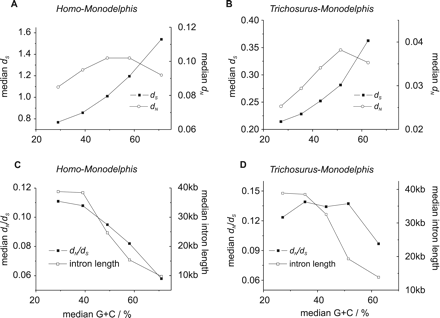
dN, dS, dN/dS, and intron length vary with G+C content at 4D sites among Monodelphis–Homo and Monodelphis–Trichosurus 1:1 orthologs. (A,C) The variation of the median values for dN, dS, dN/dS, and Monodelphis intron lengths in Monodelphis–Homo 1:1 orthologs. (B,D) The same relationships for 1:1 orthologs between the two marsupials Monodelphis and Trichosurus. The orthologs were divided into five equally populated classes according to Monodelphis G+C content at 4D sites (GC4D). We found that median dN/dS dropped from 0.12 for Trichosurus orthologs and 0.11 for Homo orthologs in the lowest G+C class (G+C <31.5% and <34.5%, respectively) to a median dN/dS of 0.10 and 0.058 in the highest G+C class (G+C >56.1% and >63.5%, respectively). The higher median dN/dS values for orthologs from the two marsupial species is likely to stem, in part, from Trichosurus EST sequencing errors that disproportionately affect dN over dS. Genes with high GC4D also exhibit higher median dS and dN values and have reduced intron lengths.











